Nat Med | A multi-omics approach to mapping the integrated tumor, immune and microbial landscape of colorectal cancer reveals the interaction of the microbiome with the immune system
Although biomarkers for primary colon cancer have been studied extensively in recent years, current clinical guidelines rely only on tumor-lymph node-metastasis staging and detection of DNA mismatch repair (MMR) defects or microsatellite instability (MSI) (in addition to standard pathology testing) to determine treatment recommendations. Researchers have noted a lack of association between gene expression-based immune responses, microbial profiles, and tumor stroma in the Cancer Genome Atlas (TCGA) colorectal cancer cohort and patient survival.
As research has progressed, quantitative characteristics of primary colorectal cancer, including cancer cellular, immune, stromal, or microbial nature of the cancer, have been reported to significantly correlate with clinical outcomes, but there is still limited understanding of how their interactions affect patient outcomes.
To dissect the relationship between phenotypic complexity and outcome, a team of researchers from the Sidra Institute of Medical Research in Qatar recently developed and validated an integrated score (mICRoScore) that identifies a group of patients with good survival rates by combining microbiome characteristics and immune rejection constants (ICR). The team performed a comprehensive genomic analysis of fresh frozen samples from 348 patients with primary colorectal cancer, including RNA sequencing of tumors and matched healthy colorectal tissue, whole exome sequencing, deep T-cell receptor and 16S bacterial rRNA gene sequencing, supplemented by whole tumor genome sequencing to further characterize the microbiome. The study was published in Nature Medicine as “An integrated tumor, immune and microbiome atlas of colon cancer”.

Article published in Nature Medicine
AC-ICAM Overview
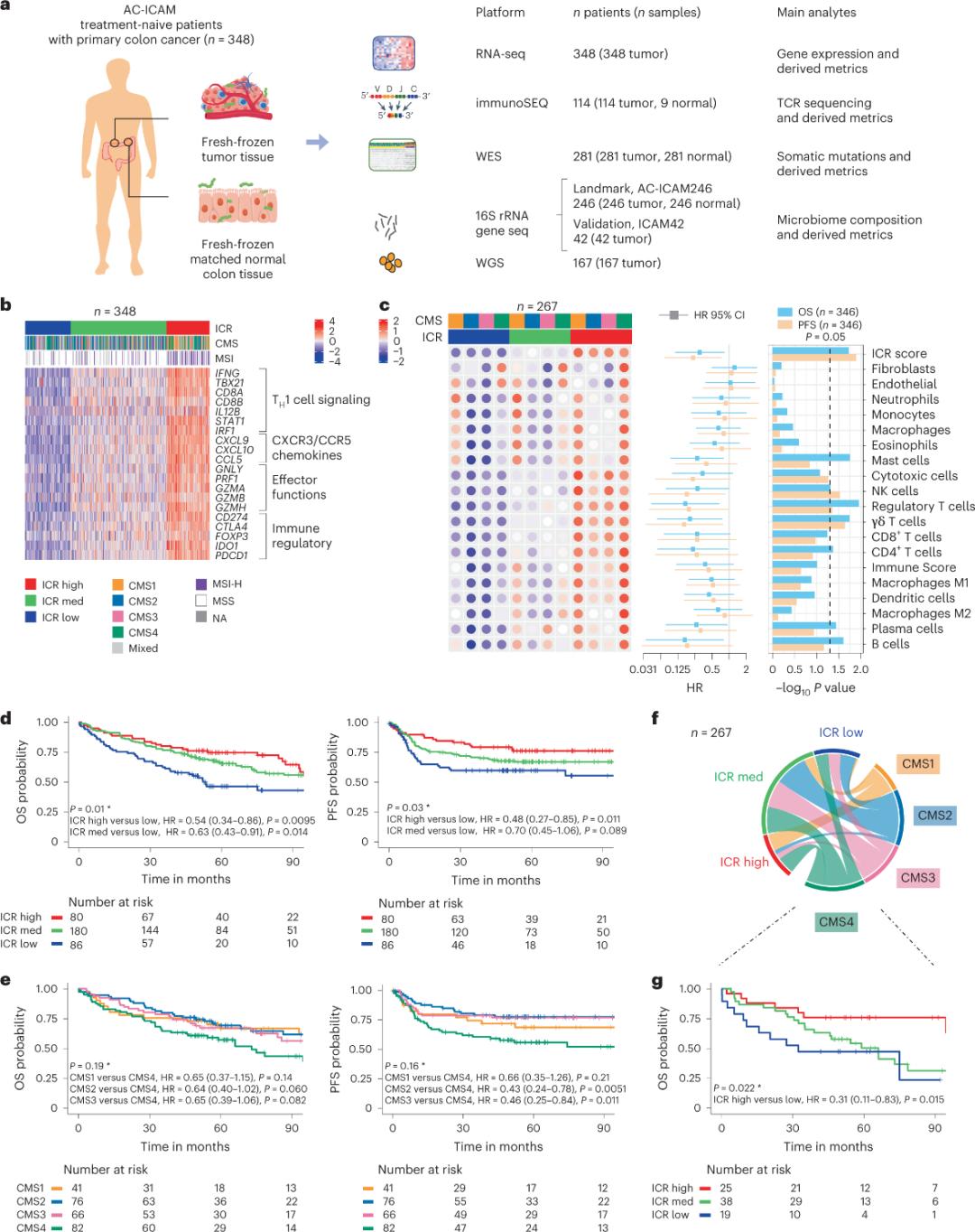
Researchers used an orthogonal genomic platform to analyze fresh frozen tumor samples and matched adjacent healthy colon tissue (tumor-normal pairs) from patients with a histologic diagnosis of colon cancer without systemic therapy. Based on whole-exome sequencing (WES), RNA-seq data quality control, and inclusion criteria screening, genomic data from 348 patients were retained and used for downstream analysis with a median follow-up of 4.6 years. The research team named this resource Sidra-LUMC AC-ICAM: A map and guide to immune-cancer-microbiome interactions (Figure 1).
Molecular classification using ICR
Capturing a modular set of immune genetic markers for continuous cancer immunosurveillance, called the immune constant of rejection (ICR), the research team optimized the ICR by condensing it into a 20-gene panel covering different cancer types, including melanoma, bladder cancer, and breast cancer. ICR has also been associated with immunotherapy response in a variety of cancer types, including breast cancer.
First, the researchers validated the ICR signature of the AC-ICAM cohort, using an ICR gene-based co-classification approach to classify the cohort into three clusters/immune subtypes: high ICR (hot tumors), medium ICR and low ICR (cold tumors) (Figure 1b). Researchers characterized the immune propensity associated with consensus molecular subtypes (CMS), a transcriptome-based classification of colon cancer. the CMS categories included CMS1/immune, CMS2/canonical, CMS3/metabolic and CMS4/mesenchymal. Analysis showed that ICR scores were negatively correlated with certain cancer cell pathways in all CMS subtypes, and positive correlations with immunosuppressive and stromal-related pathways were observed only in CMS4 tumors.
In all CMS, the abundance of natural killer (NK) cell and T cell subsets was highest in ICR high immune subtypes, with greater variability in other leukocyte subsets (Figure 1c).ICR immune subtypes had different OS and PFS, with a progressive increase in ICR from low to high (Figure 1d), validating the prognostic role of ICR in colorectal cancer.
Figure 1. AC-ICAM study design, immune-related gene signature, immune and molecular subtypes and survival.
ICR captures tumor-enriched, clonally amplified T cells
Only a minority of T cells infiltrating tumor tissue have been reported to be specific for tumor antigens (less than 10%). Therefore, the majority of intra-tumor T cells are referred to as bystander T cells (bystander T cells). The strongest correlation with the number of conventional T cells with productive TCRs was observed in stromal cell and leukocyte subpopulations (detected by RNA-seq), which can be used to estimate T cell subpopulations (Figure 2a). In the ICR clusters (overall and CMS classification), the highest clonality of immune SEQ TCRs was observed in the ICR-high and CMS subtype CMS1/immune groups (Figure 2c), with the highest proportion of ICR-high tumors. Using the whole transcriptome (18,270 genes), six ICR genes (IFNG, STAT1, IRF1, CCL5, GZMA, and CXCL10) were among the top ten genes positively associated with TCR immune SEQ clonality (Figure 2d). ImmunoSEQ TCR clonality correlated more strongly with most ICR genes than the correlations observed using tumor-responsive CD8+ markers (Figure 2f and 2g). In conclusion, the above analysis suggests that the ICR signature captures the presence of tumor-enriched, clonally amplified T cells and may explain its prognostic implications.
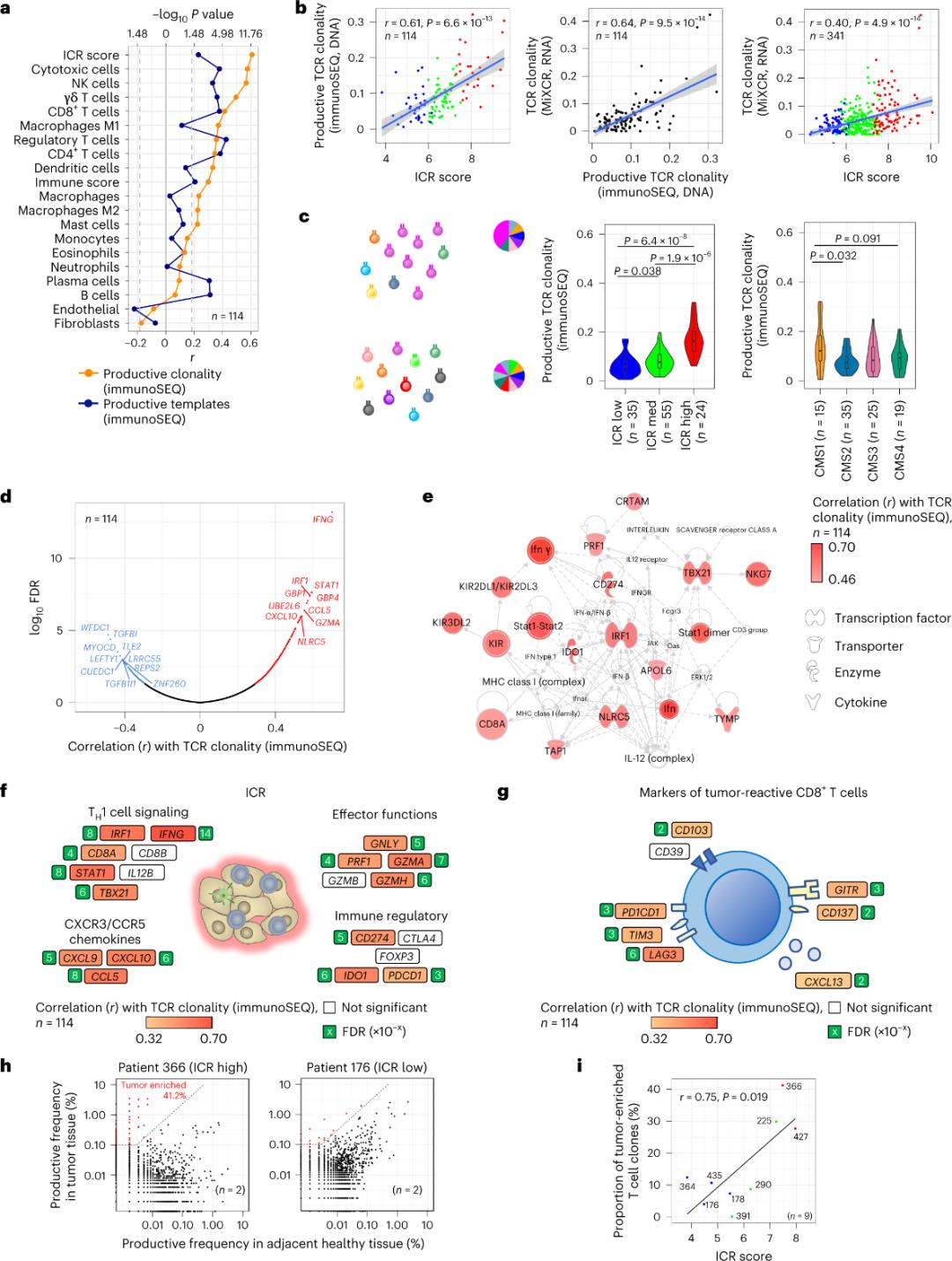
Figure 2. TCR metrics and correlation with immune-related genes, immune and molecular subtypes.
Microbiome composition in healthy and colon cancer tissues
The researchers performed 16S rRNA sequencing using DNA extracted from matched tumor and healthy colon tissue from 246 patients (Figure 3a). For validation, the researchers additionally analyzed 16S rRNA gene sequencing data from an additional 42 tumor samples that did not have matched normal DNA available for analysis. First, the researchers compared the relative abundance of flora between matched tumors and healthy colon tissue. Clostridium perfringens was significantly increased in the tumors compared to the healthy samples (Figure 3a-3d). There was no significant difference in alpha diversity (diversity and abundance of species in a single sample) between tumor and healthy samples, and a modest reduction in microbial diversity was observed in ICR-high tumors relative to ICR-low tumors.
To detect clinically relevant associations between microbial profiles and clinical outcomes, the researchers aimed to use 16S rRNA gene sequencing data to identify microbiome features that predict survival. At AC-ICAM246, the researchers ran an OS Cox regression model that selected 41 features with non-zero coefficients (associated with differential mortality risk), called MBR classifiers (Figure 3f).
In this training cohort (ICAM246), a low MBR score (MBR<0, low MBR) was associated with a significantly lower risk of death (85%). Researchers confirmed the association between low MBR (risk) and prolonged OS in two independently validated cohorts (ICAM42 and TCGA-COAD). (Figure 3) The study showed a strong correlation between endogastric cocci and MBR scores, which were similar in tumor and healthy colon tissue.
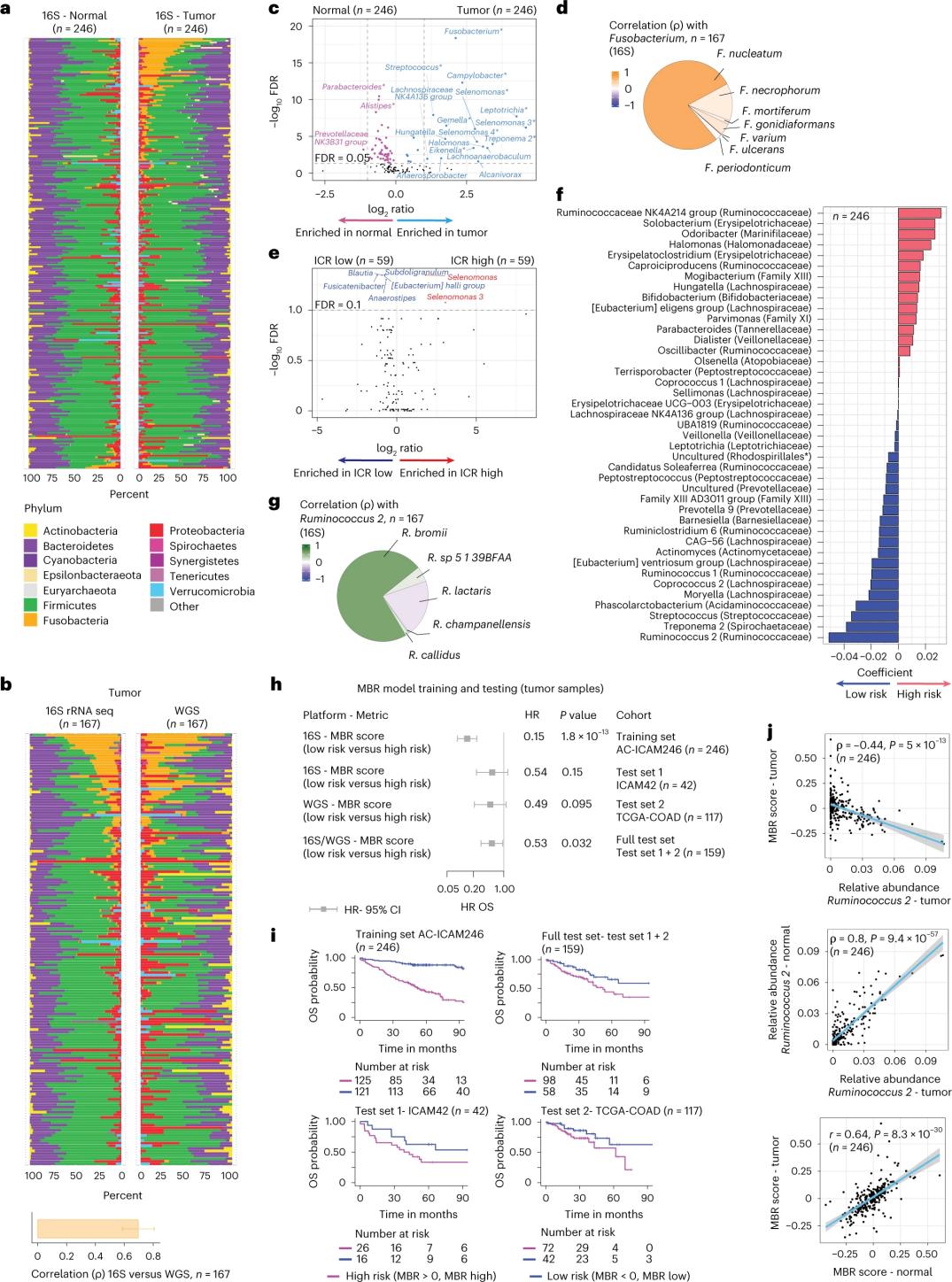
Figure 3. Microbiome in tumor and healthy tissues and the relationship with ICR and patient survival.
Conclusion
The multi-omics approach used in this study enables thorough detection and analysis of the molecular signature of the immune response in colorectal cancer and reveals the interaction between the microbiome and the immune system. Deep TCR sequencing of tumor and healthy tissues revealed that the prognostic effect of ICR may be due to its ability to capture tumor-enriched and possibly tumor antigen-specific T cell clones.
By analyzing tumor microbiome composition using 16S rRNA gene sequencing in AC-ICAM samples, the team identified a microbiome signature (MBR risk score) with strong prognostic value. Although this signature was derived from tumor samples, there was a strong correlation between healthy colorectum and tumor MBR risk score, suggesting that this signature may capture the gut microbiome composition of patients. By combining the ICR and MBR scores, it was possible to identify and validate a multi-omic student biomarker that predicts survival in patients with colon cancer. The study’s multi-omic dataset provides a resource to better understand colon cancer biology and help discover personalized therapeutic approaches.
Post time: Jun-15-2023
 中文网站
中文网站